This week we moved on from Hardy-Weinberg and focused on the process of speciation. Speciation can be defined as “reproductive isolation,” or the inability of two populations of organisms to interbreed and/or produce fertile offspring. But what would cause a population to split? And what would cause these groups to become so different they can no longer interbreed?
There are several ways this can happen. These fall into two general types of speciation: Allopatric and Sympatric speciation.

In Allopatric speciation, two populations of the same species become physically separated, usually by geographic means. This can include a river, canyon, mountain range, separating continents, etc. This is the more common type of speciation.
In Sympatric speciation, two distinct groups emerge within a population without a physical/geographic barrier between them. This type of speciation is more complicated and less common.
We also learned about the barriers of species, aka the reasons why a giraffe doesn’t mate with a pine tree. These include pre-zygotic (before fertilization) and post-zygotic (after fertilization) barriers. Mechanical isolation and gametic isolation are prezygotic because they make is mechanically impossible for organisms to mate, and for the sperm to fertilize an egg. Habitat isolation, temporal isolation and behavioral isolation are also prezygotic as they prevent organisms from even trying to mate. Postzygotic barriers include hybrid viability, reduced hybrid fertility and hybrid breakdown. These make hybrid offspring of two species either die during birth or are so unsuited to their environment they die anyways. Hybrid Breakdown indicates that the first generation of hybrid offspring are perfectly fine and viable, however after 2 or more generations severe problems begin to arise.

Questions Going Forward:
Will these species barriers relate to HWE in any way? How often do hybrid offspring become their own species? How many of the earth’s species are a product of hybrid reproduction?



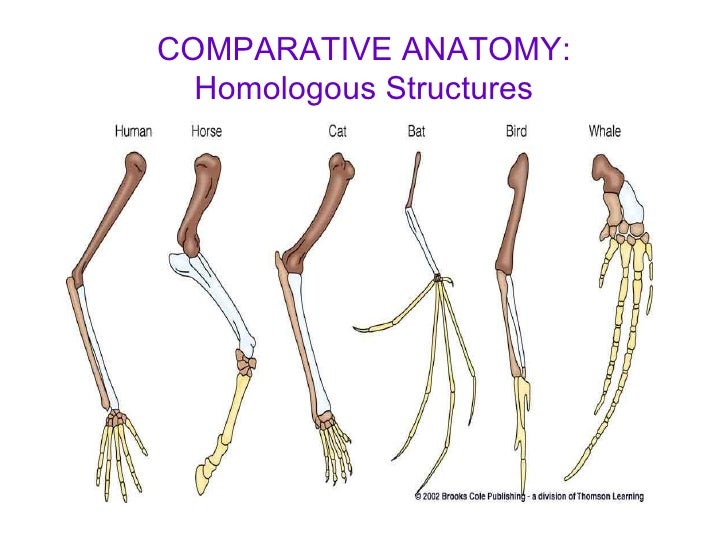
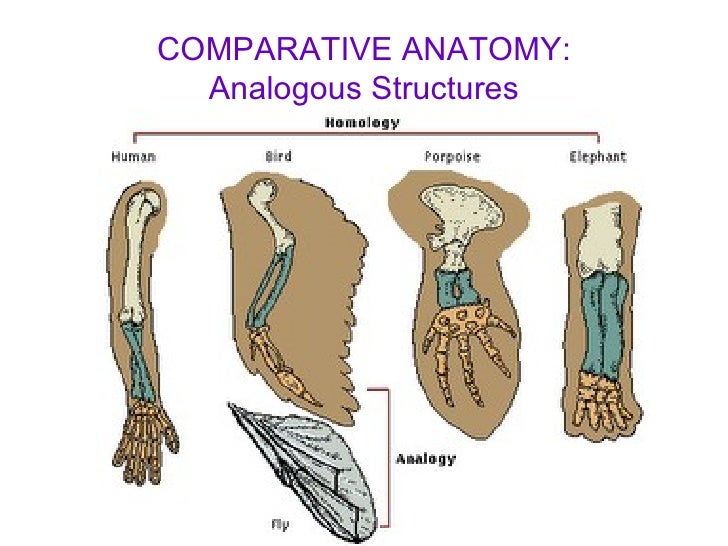 Structures: Similar adaptations to similar situations and problems (these serve as evidence for convergent evolution)
Structures: Similar adaptations to similar situations and problems (these serve as evidence for convergent evolution)